- ALL COMPUTER, ELECTRONICS AND MECHANICAL COURSES AVAILABLE…. PROJECT GUIDANCE SINCE 2004. FOR FURTHER DETAILS CALL 9443117328


Projects > ELECTRONICS > 2017 > IEEE > DIGITAL IMAGE PROCESSING
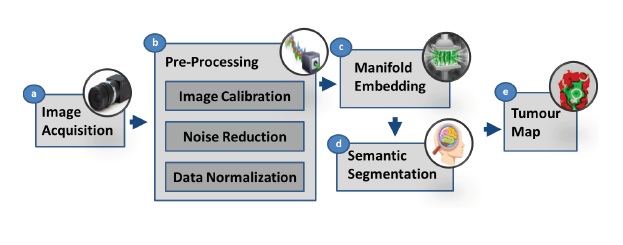
Recent advances in hyperspectral imaging have made it a promising solution for intra-operative tissue characterization, with the advantages of being non-contact, non-ionizing and non-invasive. Working with hyperspectral images in vivo, however, is not straightforward as the high dimensionality of the data makes real-time processing challenging. In this paper, a novel dimensionality reduction scheme and a new processing pipeline are introduced to obtain a detailed tumour classification map for intra-operative margin definition during brain surgery. However, existing approaches to dimensionality reduction based on manifold embedding can be time consuming and may not guarantee a consistent result, thus hindering final tissue classification. The proposed framework aims to overcome these problems through a process divided into two steps: dimensionality reduction based on an extension of the T-distributed stochastic neighbour (t-SNE) approach is first performed and then a semantic segmentation technique is applied to the embedded results by using a Semantic Texton Forest (STF) for tissue classification. Detailed in vivo validation of the proposed method has been performed to demonstrate the potential clinical value of the system.
Support Vector Machine Classification, Isomap Algorithm, Laplacian Eigenmaps and Spectral Techniques.
In this paper, we have proposed a novel manifold embedding framework FR-t-SNE with which the output generated from a hyperspectral image can be used as input for a semantic segmentation classifier of brain tissues in vivo, in situ. The proposed method aims to determine the boundaries of tumours, saving healthy brain tissue and allowing a complete resection of the malignant cells. Conventional diagnoses of internal tumours are based on excisional biopsy followed by histology or cytology. The main weaknesses of the traditional approach are twofold. Firstly, it is invasive with many potential side effects and complications; and secondly, diagnostic information is not available in real-time and requires off-line histopathology sample preparation and analysis. The proposed system can overcome these problems and tumour resection can be greatly improved during surgical procedures, thus reducing the risk of disease recurrence. Moreover, the proposed system can help in understanding the cancer progression. The real-time nature of the techniques can improve surgical accuracy, providing additional information that can also reduce the probability of erroneous resectioning of healthy tissue. The proposed framework aims to overcome these problems through a process divided into two steps: dimensionality reduction based on an extension of the T-distributed stochastic neighbour (t-SNE) approach is first performed and then a semantic segmentation technique is applied to the embedded results by using a Semantic Texton Forest (STF) for tissue classification.
BLOCK DIAGRAM